Research
Epigenetic Mechanisms of Germ Cell Development
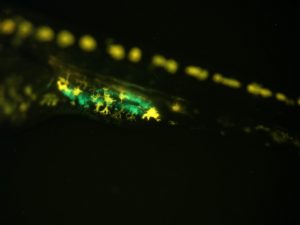
Medaka fry (female, 3 days post-hatch) with GFP in germ cells
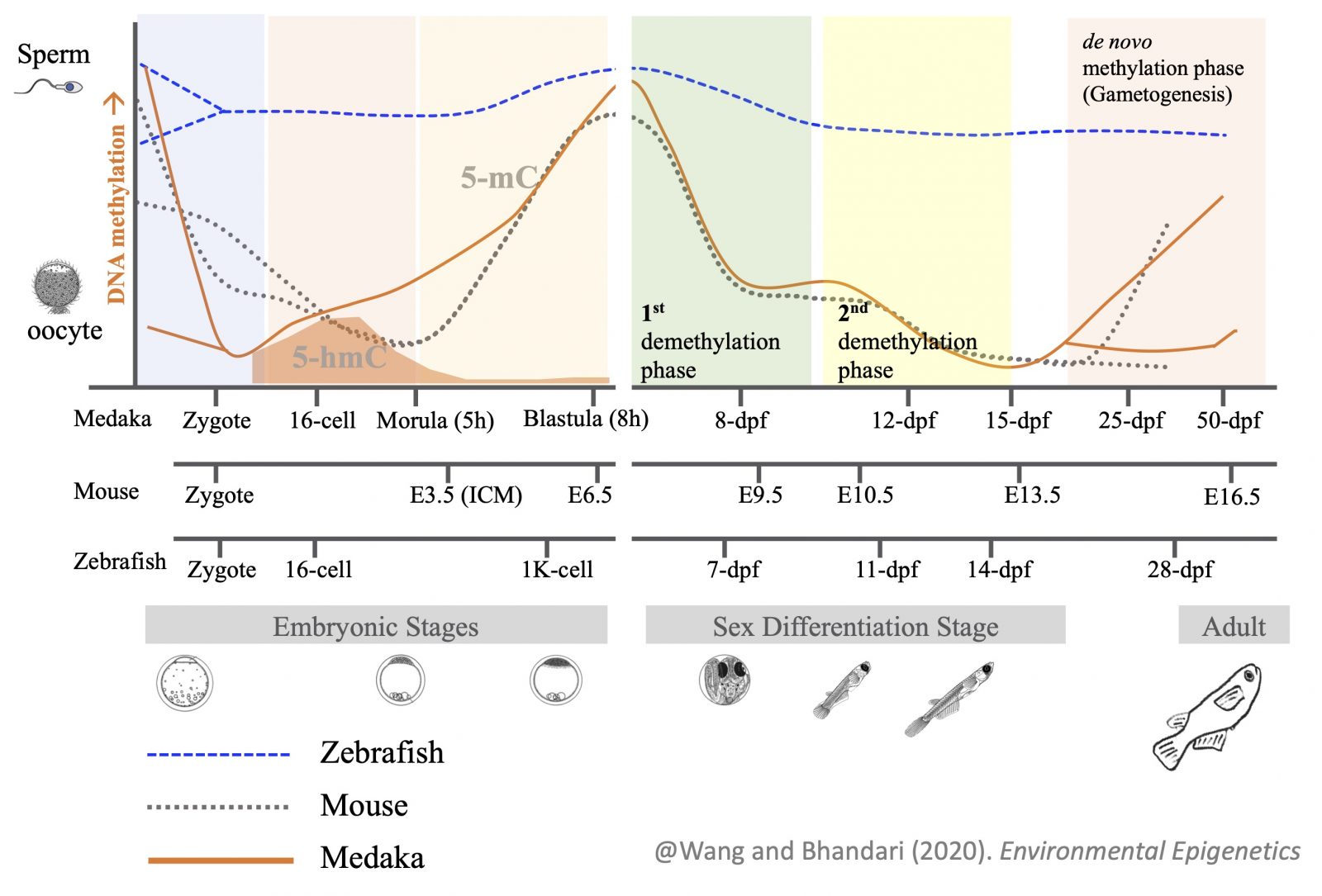
Sex differences in germ cells are believed to be epigenetically regulated, as distinct differences in DNA methylation and histone modifications have been found in germ cells during sex determination. We are studying epigenetic regulation of germ cell development, including the process of epigenetic reprogramming of primordial germ cells (PGCs), by using olvasGFP medaka as a model organism. Our research shows that medaka PGCs undergo epigenetic reprogramming (in terms of DNA methylation dynamics) in a similar fashion to mammals. We are investigating the role of the Y chromosome in epigenetic reprogramming of germ cells as fish without the Y chromosome lack this pattern of epigenetic reprogramming.
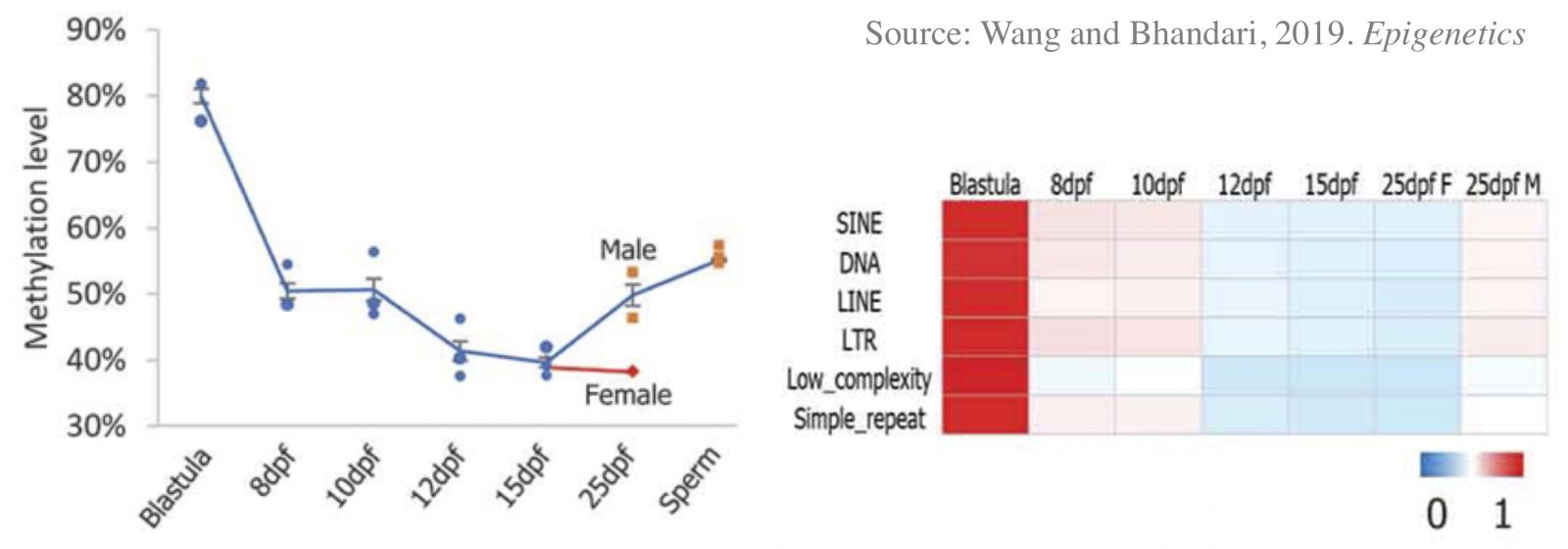
Programming of parental methylomes (genomewide DNA methylation profile) is essential for embryo development and of primordial germ cells (PGCs) for gender-specific germ cell development from its pluripotent state. Our results suggest that medaka reprogram their methylomes in early embryos and PGCs the same way mammals do. This makes medaka fish an ideal organismal model for studying epigenetic regulation of transcription in health and disease and inheritance of environmentally induced epigenetic signals.
Related publications
- Wang. X., Bhandari. R.K. (2019). DNA methylation dynamics during epigenetic reprogramming of medaka embryo. Epigenetics. 14(6):611-622. [PMID: 31010368]
- Wang, X., Bhandari, R.K. (2020). The Dynamics of DNA methylation during epigenetic reprogramming of primordial germ cells in medaka (Oryzias latipes), Epigenetics 18: 1-16 [PMID: 31851575]
- Wang, X., Bhandari, R.K. (2020). DNA methylation reprogramming in medaka fish, a promising animal model for environmental epigenetics research, Environmental Epigenetics (in press)





