The fruitfly
Drosophila ananassae


Funded by



What is the effect of natural selection on molecular variation in the genome?
One of the central questions evolutionary biologists seek to answer is how natural selection works in natural populations. Because natural selection operates on the transmission of heritable traits from one generation to the next, it ultimately traces back to the genome. We know that genomes have significant amounts of variation among individuals, populations, and species. If we can understand the evolutionary mechainsms that cause that variation to change or rearrange in the genome, we will have a detailed explanation for how evolution works in natural populations. We can then make predictions about how it worked in the past and how it might work in the future.
Drosophila has been a popular model organism for the study of evolutionary genetics for a long time. We have learned a great deal about the impact of natural selection and other evolutionary forces on genome variation during the past several decades in the genome of Drosophila melanogaster and several other related species. While at Cornell University, I started studies on particular characteristics of repetitive DNA sequences called microsatellites that are distributed across the genome of most animals. In Drosophila, the mutation rate of these sequences is several orders of magnitude lower than many other animals, including humans. We believe this may be due to a balance between a slippage mutational process during DNA replication and nucleotide substitutions that break up the repetitive sequences across the genome. Such a process appears to extend to other organisms including mice, yeast, and humans and may explain the differences in particular qualities of the repetitive sequences across the genome. Such information is critical to our understanding of the levels and patterns of variation among individuals at these sequences because we would ultimately like to use them to trace the ancestry of Drosophila populations in nature. Furthermore, our results may provide insight into the evolution of these repetitive sequences in other organisms.
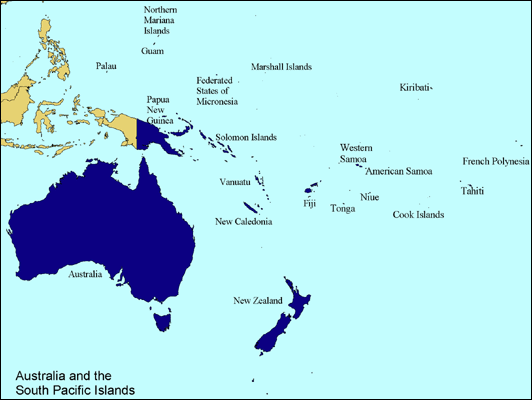
Drosophila melanogaster is cosmipolitan with geographic distributions throughout the world. Although it is an excellent organism for studies of many evolutionary questions, it will provide very little insight into how population isolation or semi-isolation affects genome evolution. Since 2002 I have been studying a species of Drosophila (D. ananassae) that is also cosmopolitan with a tropical and subtropical worldwide geographic distribution. In crontrast to D. melanogaster, populations of D. ananassae appear to be semi-isolated, creating the opportunity for local geographic adaptation. Dr. Wolfgang Stephan has found patterns of DNA variation in Asian populations that are best explained by adaptive mutations that have swept through geographically local populations. So we can use D. ananassae as a model to study the effects of limited migration among populations on molecular variation to gain insight into how natural selection and other evolutionary mechanisms work at a molecular level. With colleagues and students, I have collected population samples of D. ananassae from the South Pacific, Australia, and Asia. We are mapping the genome and studying the ancestral history of the populations and a closely related species, D. pallidosa. We have found the populations to be isolated genetically. Furthermore, they appear to be behaviorally isolated (females prefer males from their own population rather than males from others). So they may be incipient species. In addition to physiological adaptations to local geographic regions, they may also have evolved signficantly different behavioral characteristics. We are very interested in dissecting these traits genetically because they have strong impacts on fitness, a fundamental component of natural selection and are likely involved in speciation.
- Schug, M.D., S. McEvey, A. Tozier-Pearce, S.G. Smith The genetic structure of Drosophila ananassae populations from Indonesia, Australia, and the South Pacific. in review
- Moehring, A.J., J. Li, M.D. Schug, S.G. Smith, M. DeAngelis, T.F.C. Mackay, J.A. Coyne 2004. Quantitative trait loci for sexual isolation between Drosophila simulans and D. mauritiana. Genetics 167: 1265-1274.
- Schug, M.D. E.E. Regulski, A. T. Pearce, S.G. Smith 2004. Isolation and characterization of dinucleotide repeat microsatellites in Drosophila ananassae. Genetical Research 83:19-29.
- Amos, W., C.M. Hutter, M.D. Schug, C.F. Aquadro 2003. Directional evolution of size coupled with ascertainment bias for variation in Drosophila microsatellites, Molecular Biology and Evolution 20:660-662.
- Kruglyak, S., R.T. Durrett, M.D. Schug, C.F. Aquadro 2000. Distribution and abundance of microsatellites in the yeast genome can be explained by a balance between slippage events and point mutations. Molecular Biology and Evolution 17(8):1210-1219.
- Pascual M., M.D. Schug, C.F. Aquadro 2000. High density of long dinucleotide microsatellites in Drosophila subobscura. Molecular Biology and Evolution 17(8):1259-1267.
- Noor, Mohamed A.F., Malcolm D. Schug, Charles F. Aquadro. 2000. Microsatellite variation in populations of Drosophila pseudoobscura and Drosophila persimilis. Genetical Research 75:25-35.
- Schug, Malcolm D., Carolyn M. Hutter, Kris A. Wetterstrand, Mara S. Gaudette, Trudy F.C. Mackay, Charles F. Aquadro. 1998. The mutation rates of di-, tri-, and tetranucleotide repeat microsatellites in Drosophila melanogaster. Molecular Biology and Evolution 15:1751-1760.
- Hutter, Carolyn M., Malcolm D. Schug, Charles F. Aquadro. 1998. Microsatellite variation in Drosophila melanogaster and Drosophila simulans: A reciprocal test of the ascertainment bias hypothesis. Molecular Biology and Evolution 15:1620-1636.
- Schug, Malcolm D., Kris Wetterstrand, Mara S. Gaudette, Robert H. Lim, Carolyn H. Hutter, Charles F. Aquadro. 1998. The distribution and frequency of microsatellites in Drosophila melanogaster. Molecular Ecology 7:57-69.
- Schug, Malcolm D., Carolyn M. Hutter, Kris A. Wetterstrand, Mara S. Gaudette, Trudy F.C. Mackay, Charles F. Aquadro. 1998. The mutation rates of di-, tri-, and tetranucleotide repeat microsatellites in Drosophila melanogaster. Molecular Biology and Evolution 15:1751-1760.
- Hutter, Carolyn M., Malcolm D. Schug, Charles F. Aquadro. 1998. Microsatellite variation in Drosophila melanogaster and Drosophila simulans: A reciprocal test of the ascertainment bias hypothesis. Molecular Biology and Evolution 15:1620-1636.
- Schug, Malcolm D., Carolyn M. Hutter†, Mohamed A.F. Noor, Charles F. Aquadro. 1998. Mutation and evolution of microsatellites in Drosophila melanogaster. Genetica 102/103 (1/6):359-367.
- Kruglyak, Semyon, Richard Durret, Malcolm D. Schug, Charles F. Aquadro. 1998. Equilibrium distributions of microsatellite repeat length resulting from a balance between slippage events and point mutations. Proceedings of the National Academy of Sciences, USA 95:10774-10778.
- Schug, Malcolm D., Trudy F.C. Mackay, Charles F. Aquadro. 1997. Low mutation rates of microsatellite loci in Drosophila melanogaster. Nature Genetics 15:99-102.