|
Dr. Olav Rueppell |
|||||||
|
Our office and lab space are located in the Eberhart
Building on the main
UNCG campus. Click for a broad or a detailed map how to get there. Our research apiary and bee facility are located 5min to the west (see map). If you want
to DONATE directly to UNCG’s honey bee
research program, Disclaimer:
The material located at this site is not endorsed, sponsored, or provided by or on behalf of the LAST UPDATED Sept. 2011 |
Genomic Research:
Currently, we are pursuing two research problems through comparative
genomics: the exceptionally high genomic recombination rate of honey bees,
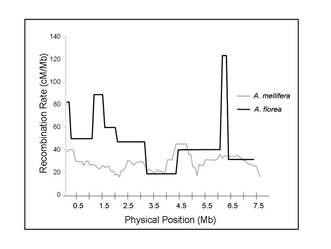
and the differences in caste divergence among various honey bees species. The average recombination rate in Apis
mellifera is about 20 cM/Mb, which is the
highest genome-wide rate reported for any metazoan. However, in collaboration
with Juergen Gadau at Arizona
State University, we have found that Apis
florea (the
red dwarf honey bee) displays a recombination rate that is at least equally
high over the two investigated chromosomes. We are interested in investigating
the evolutionary conservation of this high genomic recombination rate at
different scales, and understanding its proximate and ultimate causes. Our recent
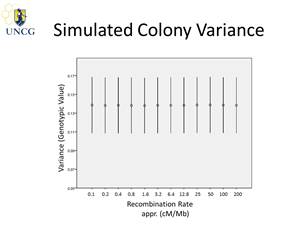
simulation studies have shown that the evolution of high recombination rates
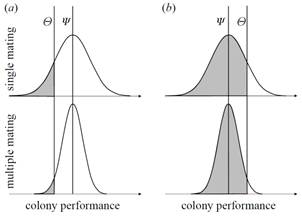
cannot be sufficiently explained by selection for increased genetic colony diversity.
On the other hand, we have shown that genetic colony diversity is generally
significant in social insects. We envision high-density linkage maps for all Apis species, which will elucidate the
patterns of recombination across different evolutionary scales and also help
the genome assemblies for these species. In addition, we are annotating genes
with putative roles in meiotic recombination in the emerging Bombus and Apis genomes in collaboration with the
Chris Elsik lab at Georgetown University.
To enable comparative genomic studies, our lab currently spearheads
the sequencing and analysis of the genome of the giant honey bee, Apis dorsata,
using only paired-end, Illumina reads in
collaboration with the Michael Schatz
(CSHL), Chris Elsik (Georgetown), and with the help of Gene Robinson (University
of Illinois). The results can be used to improve the genome predictions of
related genomes and permit comparative genomic studies in honey bees with Apis florea
and A. cerana
genomes also emerging. We are particularly interested in genes that have a
possible function in caste divergence and recombination. Furthermore, a
completed A. dorsata
genome will enable genetic mapping studies, including fine-scale analyses of
recombination.
|
||||||